Corona: Using magnetic tweezers to track down new antiviral drugs against Covid-19
How can SARS-CoV-2 be stopped? David Dulin and his team at the Max-Planck-Zentrum für Physik und Medizin in Erlangen have investigated how the virus multiplies its genome and how to impair this process at the molecular level using different active substances.
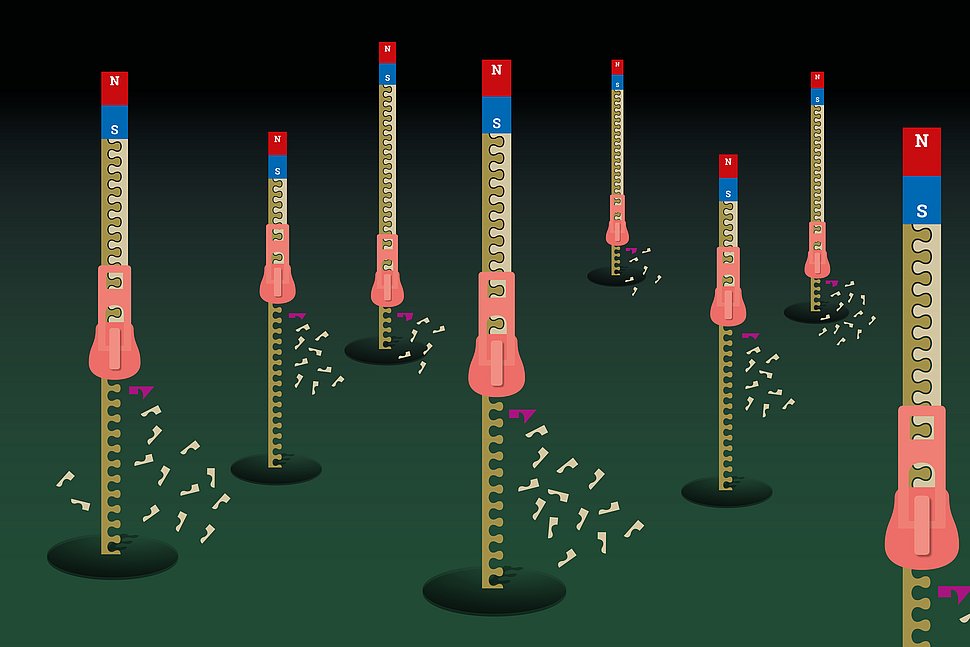
Illustration: MPZPM / Susanne Viezens
Pathogens like SARS-CoV-2 use the human cell as a factory by taking control over it and repurposing its resources to fabricate new viruses. During this process, the virus must synthesize many copies of its genome by assembling the letters of the genetic alphabet A, C, G and U - the natural nucleotides - in a specific order to form a long strand of RNA, the viral blueprint.
Because this synthesis is key to the reproduction of the virus, researchers are developing drugs to impair this process. These small molecules resemble the natural nucleotides, and are therefore called nucleotide analogues. Upon incorporation, such an agent induces either a transient or a full arrest of the RNA polymerase, the nanoscopic machine in charge of the viral genome synthesis. A well-known member of this family of drugs is remdesivir, which has been approved by the regulatory authorities of several countries for the treatment of Covid-19.
In order to rationally design nucleotide analogues, their mode of selection and incorporation in the growing RNA must be understood. To this end, a team of biophysicists led by David Dulin from VU Amsterdam, the Max-Planck-Zentrum für Physik und Medizin and the Interdisciplinary Center for Clinical Research of the Friedrich-Alexander-Universität Erlangen-Nürnberg developed a new method – based upon high-throughput magnetic tweezers – to follow single SARS-CoV-2 RNA polymerase with great precision to reveal its nucleotide addition cycle. They show that this RNA polymerase utilizes three different pathways to incorporate natural nucleotides and modulates the polymerase fidelity and viral evolution. “We now understand how the coronavirus polymerase selects and incorporates nucleotides to multiply the SARS-CoV-2 genome”, says David Dulin. They recently published these results in Cell Reports.
Rational drug design
What defines a potent nucleotide analogue is how well it is incorporated by the RNA polymerase in competition against natural nucleotides. Therefore, what is the role of the three different pathways during nucleotide analog selection and incorporation? Their research has revealed that the nature of the chemical modification in the nucleotide analog defines the pathway by which it is incorporated the growing RNA. Because these pathways have different likelihood, this defines the nucleotide analog incorporation efficiency, and therefore the antiviral efficacy. “Developing new molecules binding to the RNA polymerase to increase the proportion of a given pathway is a potential route to improve nucleotide analogue therapeutic efficacy”, says one of the co-authorsm, Martin Depken of the TU Delft.
These results obtained by the group of David Dulin are now published in a second article in the journal eLife. "It will take some time, possibly years, for these observations to yield a therapeutic to treat Covid-19", explains one of the co-authors Craig Cameron, a virologist at UNC Chapel Hill, and continues: “So far, we have only repurposed antiviral drugs against other viruses. We simply have no understanding of what makes a given nucleotide analogue a good substrate for the coronavirus polymerase. We now need to rationally design the next generation of nucleotide analogues”. The work of David Dulin is a step towards this goal.
Original publications:
"The nucleotide addition cycle of the SARS-CoV-2 polymerase", Cell Reports, doi.org/10.1016/j.celrep.2021.109650
"Inhibition of SARS-CoV-2 polymerase by nucleotide analogs from a single-molecule perspective", eLife, doi.org/10.7554/eLife.70968
Contact:
David Dulin
Contact
Edda Fischer
Head of Communication and Marketing
Telefon: 09131 7133 805
MPLpresse@mpl.mpg.de






